library(igraph)
edges = c("1", "2",
"2", "3",
"1", "3")
g = make_graph(edges = edges, directed = FALSE)
E(g)$label = c(expression(a[1]), expression(a[2]), expression(a[3]))
plot(g)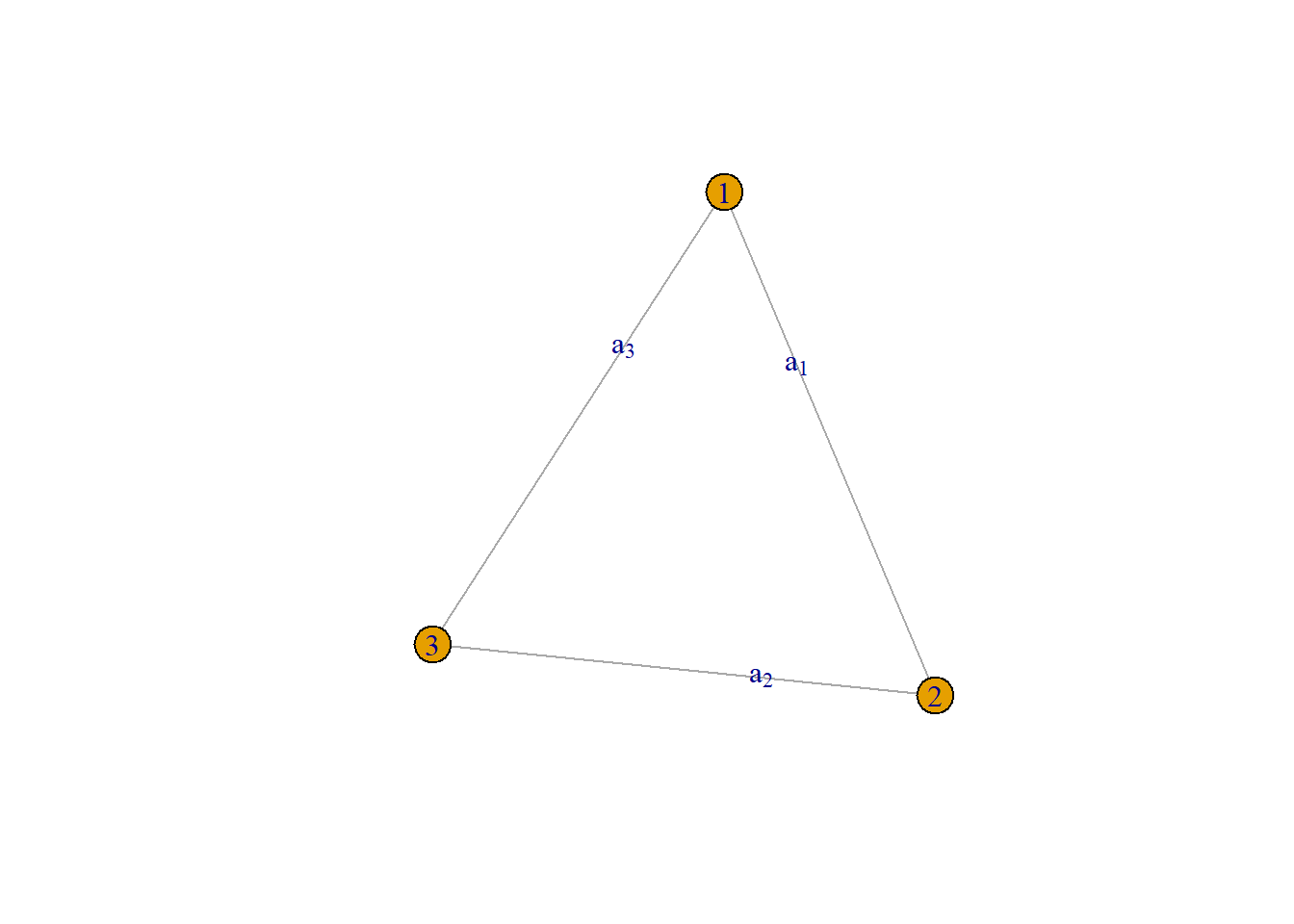
In graph theory, adjacency matrices are typically square matrices which are used to represent graphs. The elements of an adjacency matrix denote whether a set of vertices are linked or not. This means that the “cells” of an adjacency matrix are \(0.00\) when two vertices are not linked and \(1.00\) when they are linked.
There are cases when the links can be different from \(0.00\) or \(1.00\) and we’ll cover those next week.
Let’s recall the original structure of our simple undirected graph:
\[G = (V,E)\]
where:
Recall, this basic graph:
If we translate this to a matrix form, we get the following adjacency matrix:
\[ \underset{3\times 3}{\mathbf{A}} = \begin{bmatrix} 0 & a_{1} & a_{2} \\ a_{1} & 0 & a_{3} \\ a_{2} & a_{3} & 0 \end{bmatrix} \]
Notice that the diagonal elements are all 0. This aligns with the first argument that we cannot have self-loops. Also notice that certain cells have the same names. This is because each link between vertices can only happen once and is undirected.
Let’s try creating the graph defined by this adjacency matrix:
library(igraph)
edges = c("1", "2",
"2", "3",
"1", "3")
g = make_graph(edges = edges, directed = FALSE)
E(g)$label = c(expression(a[1]), expression(a[2]), expression(a[3]))
plot(g)
See that the diagonal elements of an undirected graph is \(0.00\) because it forbids self-loops. Likewise, there are only 3-links aligning with our adjacency matrix.
Now, what about our digraph from earlier? What would the matrix of that look like? If we had the following defintion for our edges:
\[E \subseteq \{(x, y) \mid x, y \in V\}\]
This then generates the following adjacency matrix:
\[ \underset{3\times 3}{\mathbf{A}} = \begin{bmatrix} a_{1} & a_{2} & a_{3} \\ a_{4} & a_{5} & a_{6} \\ a_{7} & a_{8} & a_{9} \end{bmatrix} \] Notice that the diagonal elements are no longer \(0.00\) and the off-diagonal elements no longer match.
We can then create the resulting graph implied by this adjacency matrix:
edges = c("1", "1",
"1", "2",
"1", "3",
"2", "1",
"2", "2",
"2", "3",
"3", "1",
"3", "2",
"3", "3")
g = make_graph(edges = edges, directed = TRUE)
E(g)$label = c(expression(a[1]), expression(a[2]), expression(a[3]),
expression(a[4]), expression(a[5]), expression(a[6]),
expression(a[7]), expression(a[8]), expression(a[9]))
E(g)$curved = -0.3
plot(g, edge.arrow.size = 0.5)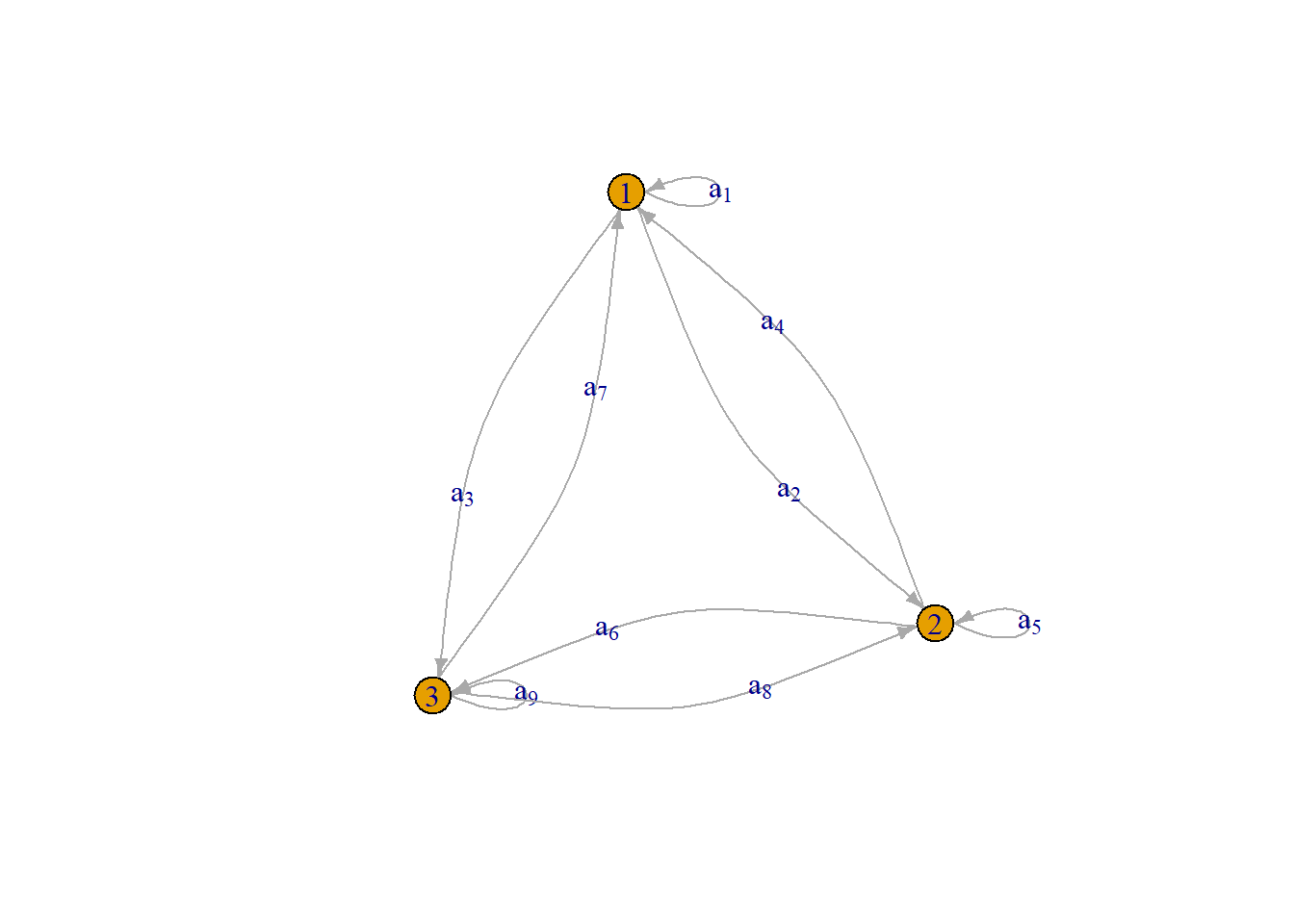
Notice that–unlike our simple undirected graph–this has 2 sets of links between each vertex pair and they have arrows denoting the direction of the association.
What networks have you seen or worked with that are digraphs? Perhaps more challenging, what networks do you think are more appropriate as undirected rather than directed graphs? Why?
We end today’s meeting with a discussion on matrix exponents and the adjadency matrix. We already learned that for matrices to be multiplied against one another they must be “conformable”.
When we are exponentiating a matrix, conformability is not an issue because they are identical and therefore conformable.
An interesting property arises when exponentiating an adjacency matrix which is that \(\underset{n\times n}{\mathbf{A}}^k\) produces a matrix where each cell denotes the number of \(^{k}\) length paths or “walks” exist between two vertices. Let’s illustrate what that means with a simple example of a squared adjacency matrix:
\[ \underset{n\times n}{\mathbf{A}^{2}} = \begin{bmatrix} a_{11} & a_{12} & a_{13} \\ a_{21} & a_{22} & a_{23} \\ a_{31} & a_{32} & a_{33} \end{bmatrix} \] Here, the cell \(a_{21}\) would represent the number of 2-step paths that connect the two vertices in question. We can illustrate this more concretely with a larger graph:
set.seed(8391)
g = sample_pa(n = 10,
power = 1.0,
m = 2,
directed = FALSE)
plot(g)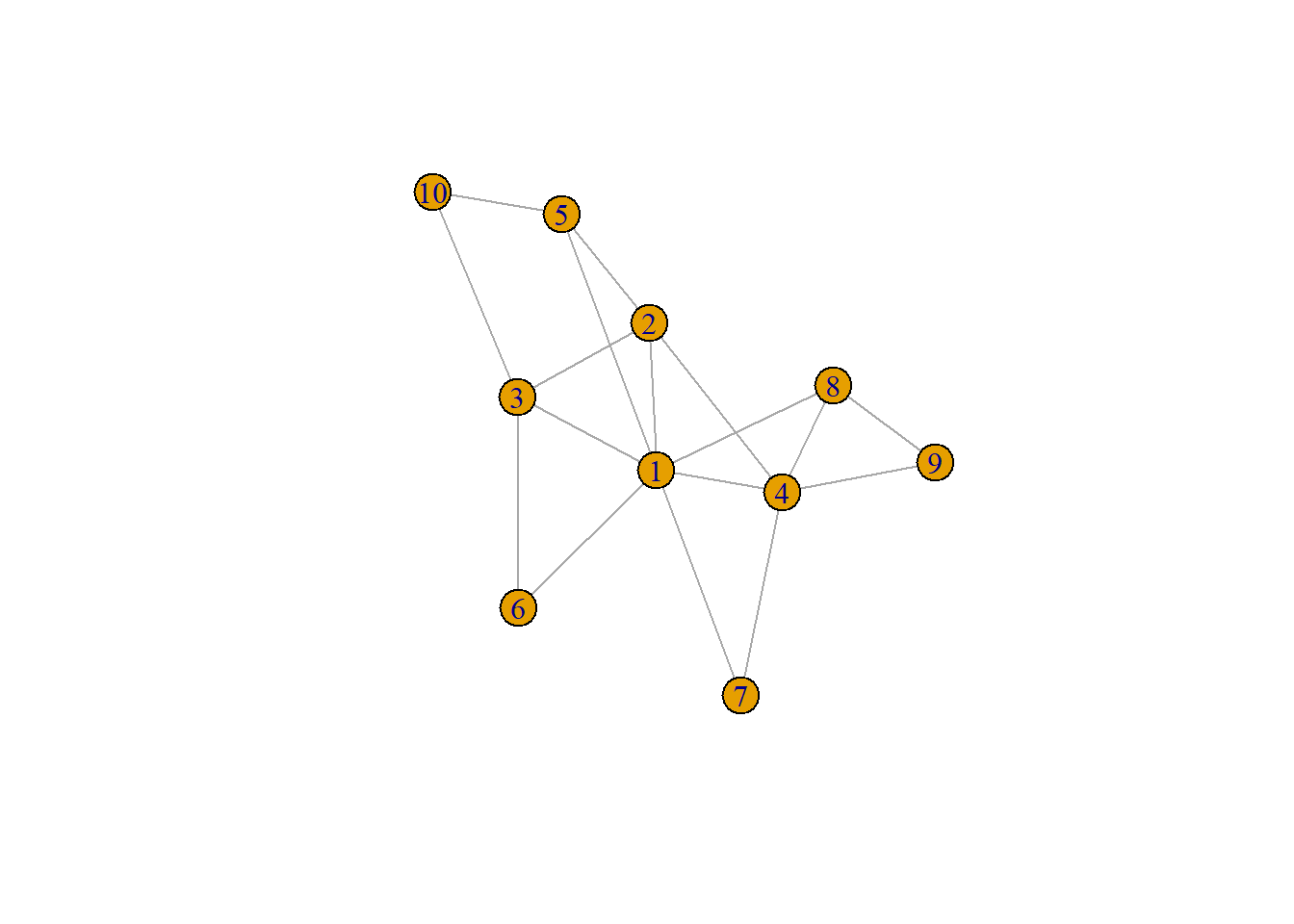
adj_matrix = as_adjacency_matrix(g, sparse = FALSE)
adj_matrix %*% adj_matrix [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10]
[1,] 7 3 2 3 1 1 1 1 2 2
[2,] 3 4 1 1 1 2 2 2 1 2
[3,] 2 1 4 2 3 1 1 1 0 0
[4,] 3 1 2 5 2 1 1 2 1 0
[5,] 1 1 3 2 3 1 1 1 0 0
[6,] 1 2 1 1 1 2 1 1 0 1
[7,] 1 2 1 1 1 1 2 2 1 0
[8,] 1 2 1 2 1 1 2 3 1 0
[9,] 2 1 0 1 0 0 1 1 2 0
[10,] 2 2 0 0 0 1 0 0 0 2Finally, a minor challenge:
Some graphs have properties that are unlike those we covered today. One such graph is the bipartite graph which–in its simplest form–is a graph composed of two sets of vertices.
For example, we may have \(V_{1}\) which is a set of researchers and \(V_{2}\) which is a set of publications where \(V_{1} \cap V_{2} = \emptyset\). For regular humans, this translates to: The intersection of \(V_{1}\) and \(V_{2}\) contains no elements; that is, they share no parts.
\(E\subseteq \{\{x, y\} \mid x \in V_{1}, y\in V_{2}\}\)
We may forgo \(x\neq y\) because \(x\) and \(y\) are in separate sets therefore they cannot be equal given the definition: \(V_{1} \cap V_{2} = \emptyset\)